Contents
clear all
close all
targetDIR = 'Data';
User Input Variables
goose = 1.0;
Pix2um = 0.82;
StainType = 1;
statement = ['cd ' targetDIR];
eval(statement)
imsDIR = dir('*.TIF');
nIMS = length(imsDIR);
holes = 0;
if ~isdir('OutPut')
mkdir('OutPut')
end
cd OutPut
d = {'Data Name','Mean Epi Thickness','SD Epi Thickness'};
xlswrite('OutPut.xls', d, 1, 'A1')
cd ..
cycle through all images to find epidermis
for i_im = 1:nIMS
try
tic
disp(sprintf('Now Crunching %4.0f of %4.0f',i_im,nIMS))
current_name = imsDIR(i_im).name;
clear I Ny Nx Nz blue green red threshold bw bw2 bw3 bw4 bw5 bw6 B L stats bw7 bw8 val Y_crd y_coords
I = double(imread(current_name));
[Ny Nx Nz] = size(I);
blue = zeros(Ny,Nx);
green = zeros(Ny,Nx);
red = zeros(Ny,Nx);
blue = I(:,:,1);
green = I(:,:,2);
red = I(:,:,3);
threshold = graythresh(green/max(max(green)));
bw = im2bw(green/max(max(green)), threshold*goose);
if StainType
ratioRED = red./blue;
threshold = graythresh(ratioRED/max(max(ratioRED)))*goose;
bw = im2bw(ratioRED/max(max(ratioRED)), threshold);
end
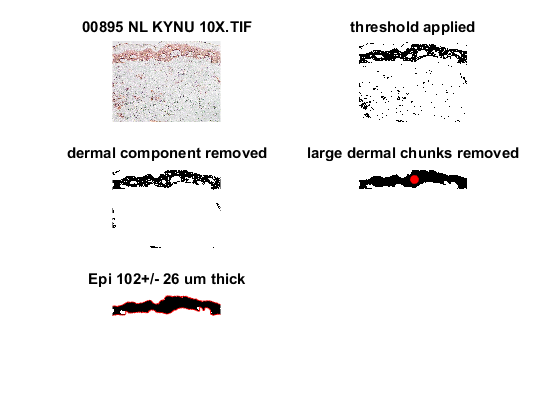
figure(1);clf
subplot(3,2,1)
imagesc(I/max(max(max(I))))
title(current_name)
axis equal image
axis off
subplot(3,2,2)
imagesc(bw);
colormap gray
axis equal image
axis off
title('threshold applied')
bw2 = imfill(bw,'holes');
subplot(3,2,3)
imagesc(bw2);
hold on
axis equal image
axis off
title('dermal component removed')
bw3 = 1-bw2;
bw4 = imfill(bw3,'holes');
seD = strel('diamond',1);
bw5 = imerode(bw4,seD);
bw6 = bwareaopen(bw5,5000);
[B, L] = bwboundaries(bw6,'nohole');
stats = regionprops(L,'Centroid','PixelList');
bw7 = 1-bw6;
subplot(3,2,4)
imagesc(bw7);
hold on
axis equal image
axis off
bw8 = bw7;
for iCTR = 1:length(stats)
y_coords(iCTR) = stats(iCTR).Centroid(2);
plot(stats(iCTR).Centroid(1),stats(iCTR).Centroid(2),'ro','markerfacecolor','r')
end
[val Y_crd] = min(y_coords);
for iCTR = 1:length(stats)
if stats(iCTR).Centroid(2) > 1.4* stats(Y_crd).Centroid(2)
cor_coords = stats(iCTR).PixelList;
for i_corect = 1:length(cor_coords)
bw8(cor_coords(i_corect,2),cor_coords(i_corect,1)) = 1;
end
end
end
title('large dermal chunks removed')
axis equal image
axis off
drawnow
subplot(3,2,5)
imagesc(bw8);
hold on
axis equal image
axis off
ind_epi = 0;
for ix = 1:Nx
iy = 1;
while bw8(iy,ix) > 0
iy = iy + 1;
if iy == Ny
bw8(iy,ix) = 0;
end
end
top(ix) = iy;
iy = Ny;
while bw8(iy,ix) > 0
iy = iy - 1;
if iy == 1
bw8(iy,ix) = 0;
end
end
bottom(ix) = iy;
if bottom(ix) ~= top(ix)
ind_epi = ind_epi+1;
count_white = 0;
iy = top(ix);
while iy < bottom(ix)
iy = iy + 1;
if bw8(iy,ix) == 1
count_white = count_white + 1;
end
end
true_thick(ind_epi) = bottom(ind_epi) - top(ind_epi) - count_white;
end
end
plot([1:Nx],top,'r-')
plot([1:Nx],bottom,'r-')
epithick = bottom - top;
title(sprintf('Epi %3.0f+/-%3.0f um thick',mean(true_thick)*Pix2um,std(true_thick)*Pix2um))
name = sprintf(['Out_Put', current_name]);
if ~isdir('OutPut')
mkdir('OutPut')
end
cd OutPut
print(figure(1),'-dtiff','-r600',name);
clear d
d = {current_name, mean(true_thick)*Pix2um, std(true_thick)*Pix2um};
xlswrite('OutPut.xls', d, 1, sprintf('A%1.0f',i_im+1))
cd ..
toc
catch
end
end
Now Crunching 1 of 2
Elapsed time is 9.498498 seconds.
Now Crunching 2 of 2
Elapsed time is 8.778257 seconds.